The Sanderson Group Webpages
Department of Chemistry
Durham University, Durham, UK
Amino Acid-Lipid Interactions
A large number peptides and proteins are capable of insertion into lipid membranes, examples of which include signal sequences, antimicrobial peptides and viral coat proteins. Peripheral membrane proteins, on the other hand, bind to the membrane surface without insertion. Of fundamental interest to us are the underlying mechanisms of protein binding to the membrane surface, and the factors that determine their ability to insert into the bilayer. Allied to this is an understanding of the stabilising interations that exist between integral membrane proteins and their lipid environment. Specific amino acids appear to have key roles in these processes.
Modelling the Interactions of Peptides with Lipids
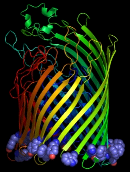
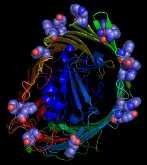
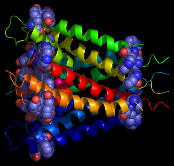
The important lipid-anchoring role played by aromatic residues in membrane proteins, particularly those located in the transition zone between the low dielectric lipid interior and the polar lipid head groups (Figure 1), is well established.1,2
Figure 1
From left to right: aromatic resdues clustered around the interior surface of the
Ferrichrome-Iron Receptor Precursor (1by3.pdb), a prokaryotic outer
membrane protein involved in ferric ion transport, and the possium
channel (KCSA) from Streptomyces lividans (1bl8.pdb). In the case of the latter, all of the
aromatic residues in the protein are highlighted, and the tendency to
cluster in the region of the membrane interface region is evident.
The peptide backbones are shown as ribbons, and the aromatic residues
as spacefilled Van der Waals spheres.
This has been confirmed by our own bioinformatics exercises on the relatively few membrane proteins in the PDB. In order to model the interactions between aromatic amino acids and lipids more extensively, we are in the process of developing simple models (see below) which will provide us with accurate structural and thermodynamic data for a range of amino acid-lipid combinations. We aim ultimately to be able to synthesise non-natural amino acids that will produce enhanced peptide interactions with lipid membranes.
A Model System for the Study of Peptide-Lipid Interactions
We have employed a simple system1,2 to test the hypothesis that aromatic amino acids have interactions with the headgroups of neutral lipids such as phosphatidylcholines that are significantly different from other amino acids. The system (Figure 2) is composed of interacting pairs of molecules that are soluble in non-competitive solvents such as chloroform whilst retaining all of the chemical functionality present in the natural polymers.
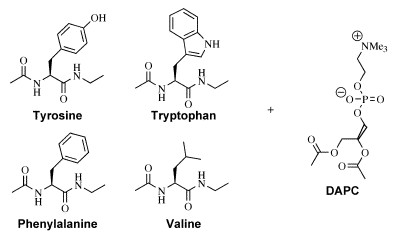
Figure 2 Host-Guest systems used to
study peptide-lipid interactions.
Each of the amino acid derivatives indicated is suitable for use as a host in an NMR titration
using the lipid analogue DAPC as guest.
When Diacetyl
Phosphatidylcholine (DAPC) is used as a guest for binding
studies (by NMR) with each of the amino acids in turn, both the
Tryptophan and Tyrosine derivatives exhibit interactions with DAPC that
are significantly stronger than either the Phenylalanine or Valine
derivatives. The latter two amino acids have association
constants that are the same (within experimental error), indicating
that Phenylalanine has little preference for interaction with the lipid
headgroup compared with an aliphatic amino acid such as Valine. In the
case of the Tryptophan and Tyrosine derivatives, both preferentially
form 2:1 adducts with DAPC (with macroscopic association constants >4000 M-1)
that result in significant complexation-induced chemical shift changes.
In the case of the Tryptophan:DAPC adduct, cross-peaks arising form
intermolecular interactions can be observed in the 2D ROESY spectrum of
a mixture of the two. We have used these data to generate models of the
Tryptophan:DAPC adduct by a combination of simulated annealing protocols
(Figure 3) and semi-empirical modelling.
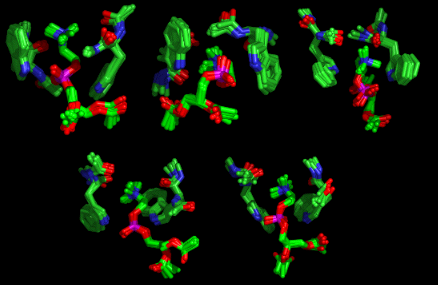
Figure 3 Structures of the Tryptophan:DAPC adduct generated by Simulated Annealing

Although a large
number of models are produced, consistent with the dynamic nature of the intermolecular
interactions, there are nevertheless some features common to all of the
structures:
- Cation-π interactions
- Indole-NH---phosphate hydrogen bonds
- Cation-carbonyl coordination
- Amide-NH---phosphate hydrogen bonds
Trends in the Free Energy of Association of Tryptophan Analogues with PC
More recently we have examined electrostatic effects on the interactions between tryptophan and phosphocholines. In this work, the lipid DMPC was used as the guest in NMR titrations with 5- and 6-substituted tryptophan analogues.3 The data were modelled to include association with water and lipid micellisation. The results indicate that association of tryptophan with phosphocholines under ideal conditions follows an "inverse-U" relationship with Hammett parameter of the indole substituent. Tryptophan (R=H) produced the weakest-associating complexes. The addition of electron-releasing (R=OMe) or electron-withdrawing (R=NO2) groups to the indole ring both produced more favourable free energies of association with the lipid. These trends have been accounted for in terms of increased hydrogen bond donor ability of the indole N-H for electron withdrawing groups, and stronger cation-π interactions when the indole substituent is electron releasing.
References
- "Characterisation
of the
Interactions of
Aromatic Amino Acids with Diacetyl Phosphatidylcholine", John M.
Sanderson and Eleanor J. Whelan, PCCP,
2004, 6, 1012-1017
- "Peptide-Lipid
Interactions: Insights and Perspectives", John M. Sanderson Org. Biomol. Chem., 2005, 3, 201-212.
- "Free-Energy Relationships for the Interactions of Tryptophan with Phosphocholines", Georg Blaser, John M. Sanderson and Mark R. Wilson, Org. Biomol. Chem., 2009, 7, 5119-5128.
Downloads
Download
spreadsheets for curve-fitting of NMR titration data here